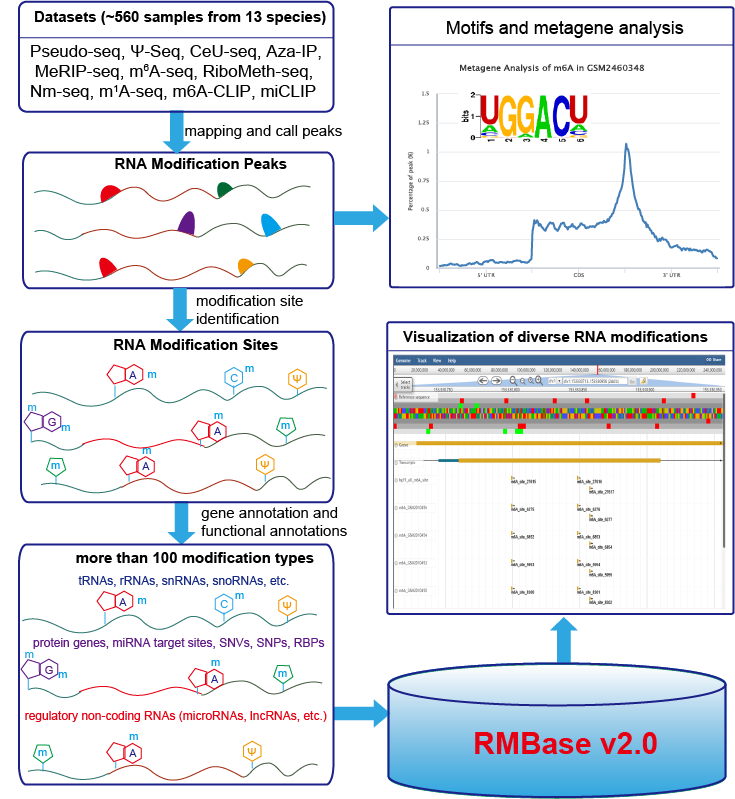
RNA Modification Database, RMBase v2.0: deciphering the map of RNA modifications from epitranscriptome sequencing data (Pseudo-seq, Ψ-seq, CeU-seq, Aza-IP, MeRIP-seq, m6A-seq, RiboMeth-seq, m1A-seq).

Frontiers | Emerging Perspectives of RNA N6-methyladenosine (m6A) Modification on Immunity and Autoimmune Diseases

Figure 2 from High-resolution N(6) -methyladenosine (m(6) A) map using photo-crosslinking-assisted m(6) A sequencing. | Semantic Scholar

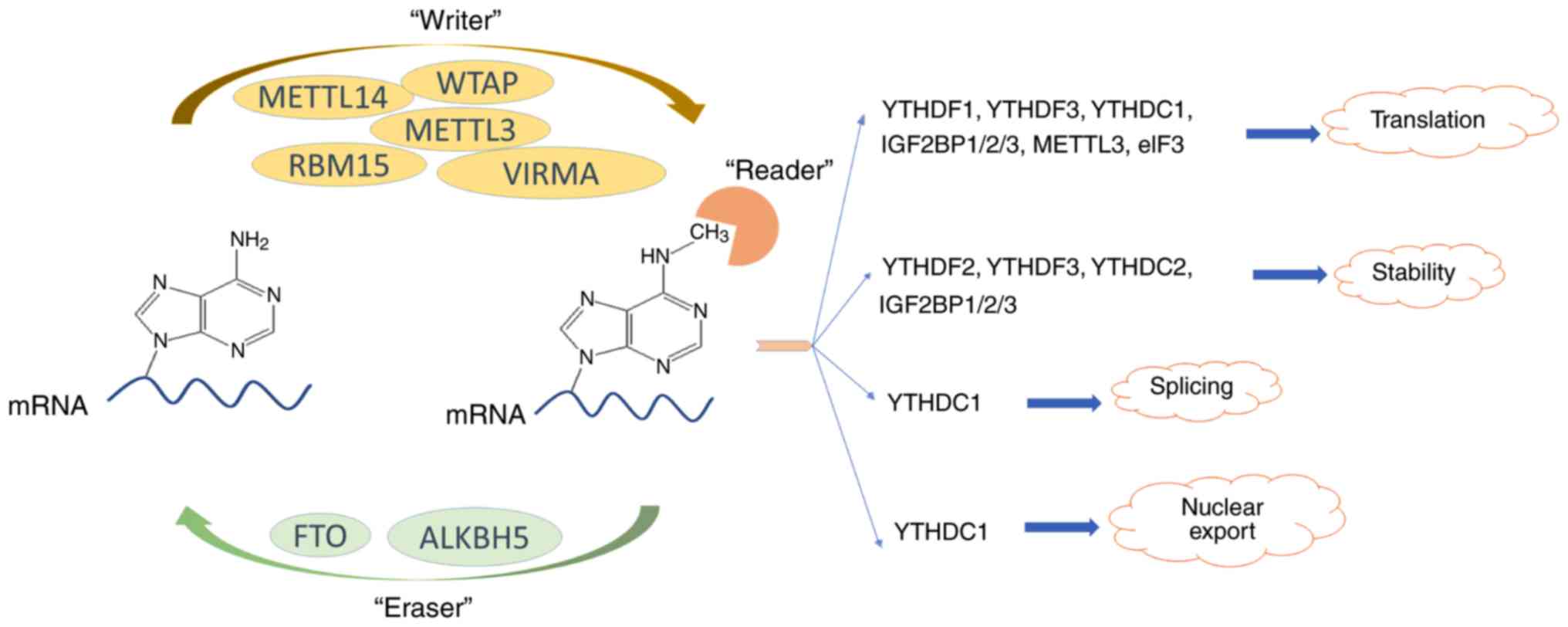
The role of m6A modification in the biological functions and diseases | Signal Transduction and Targeted Therapy

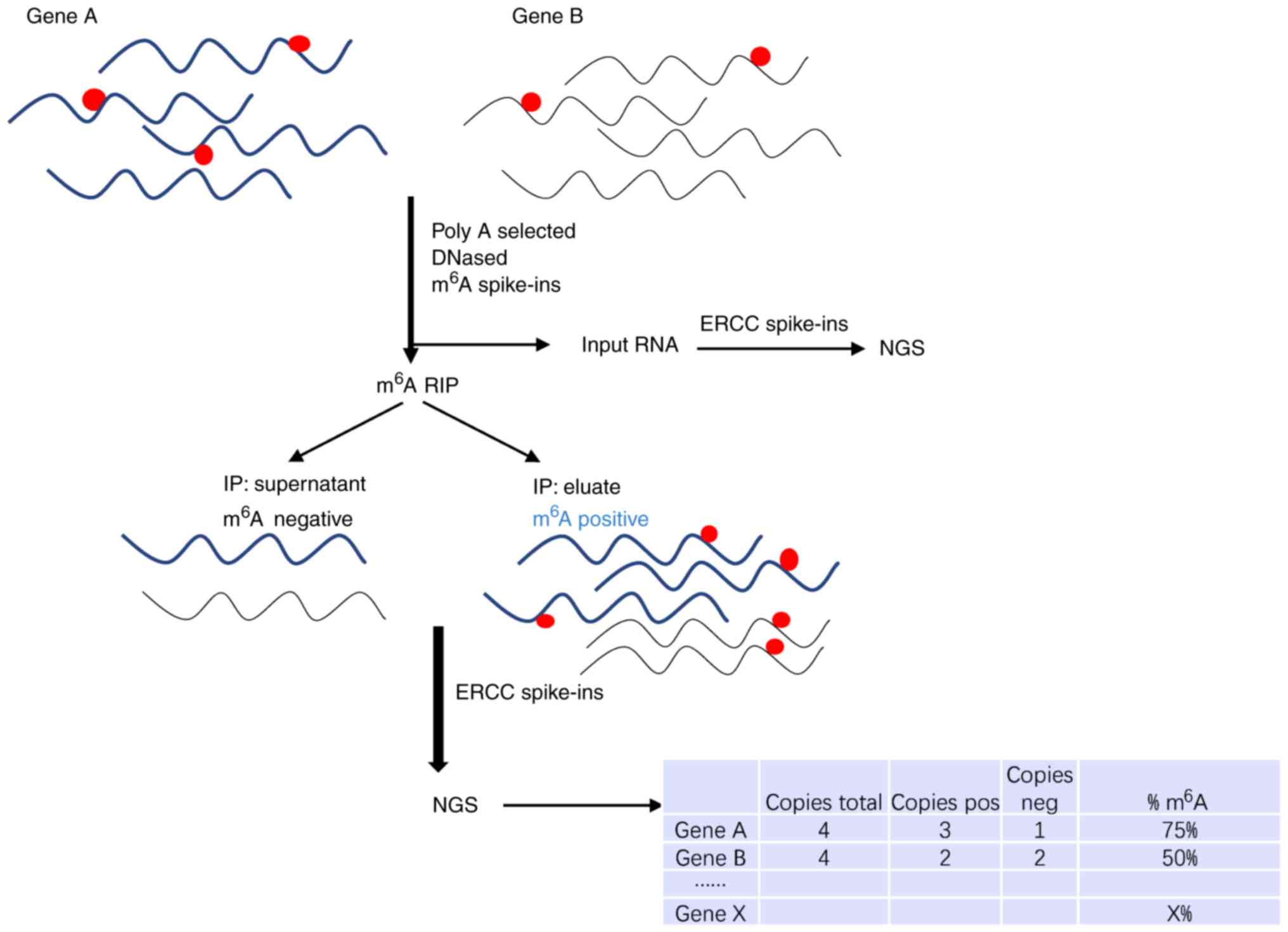
Transcriptome-wide mapping of N6-methyladenosine by m6A-seq based on immunocapturing and massively parallel sequencing | Nature Protocols

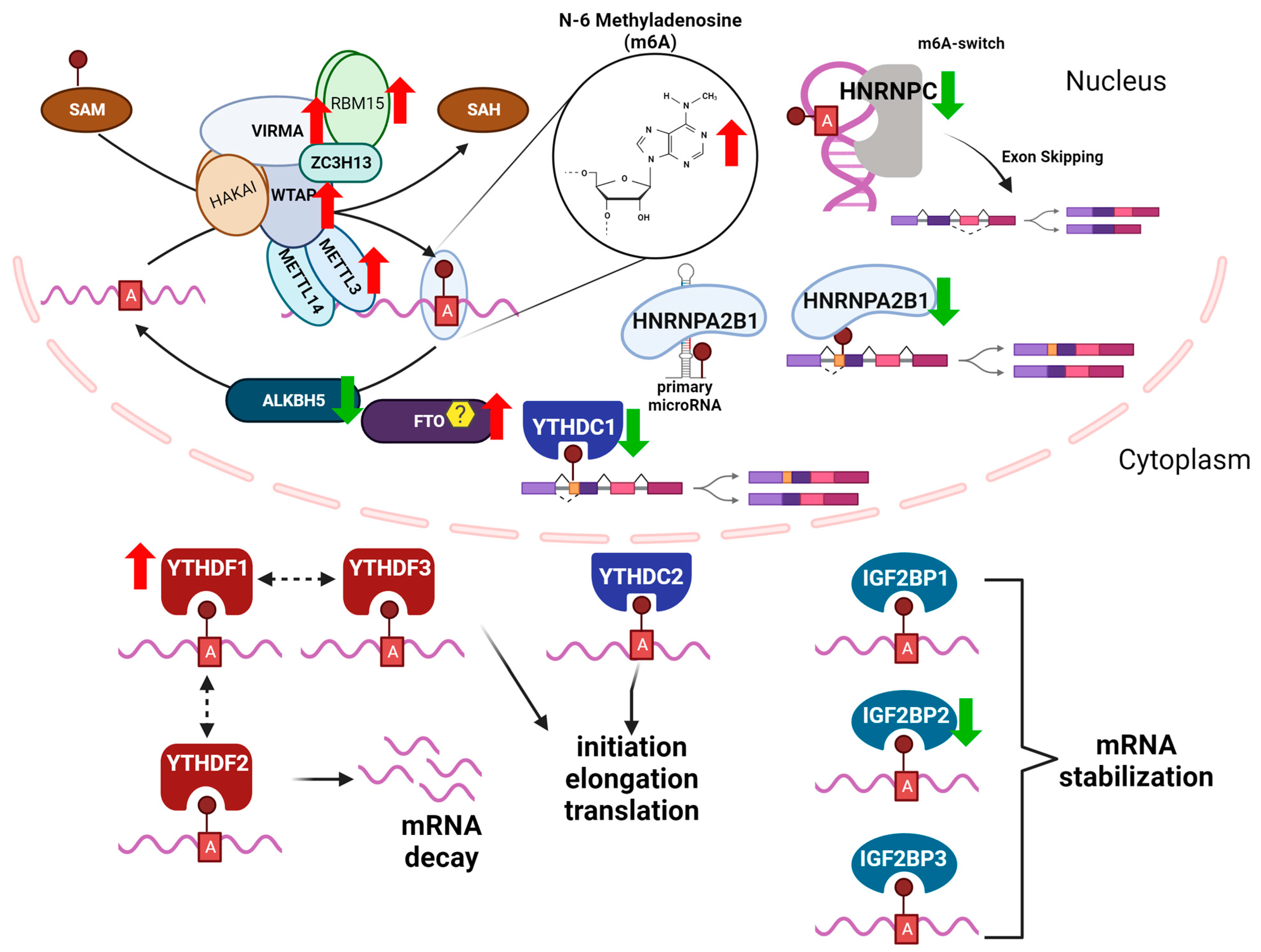
Relevance of N6-methyladenosine regulators for transcriptome: Implications for development and the cardiovascular system - Journal of Molecular and Cellular Cardiology

Single-nucleotide resolution achieved by m 6 A-CLIP/immunoprecipitation... | Download Scientific Diagram
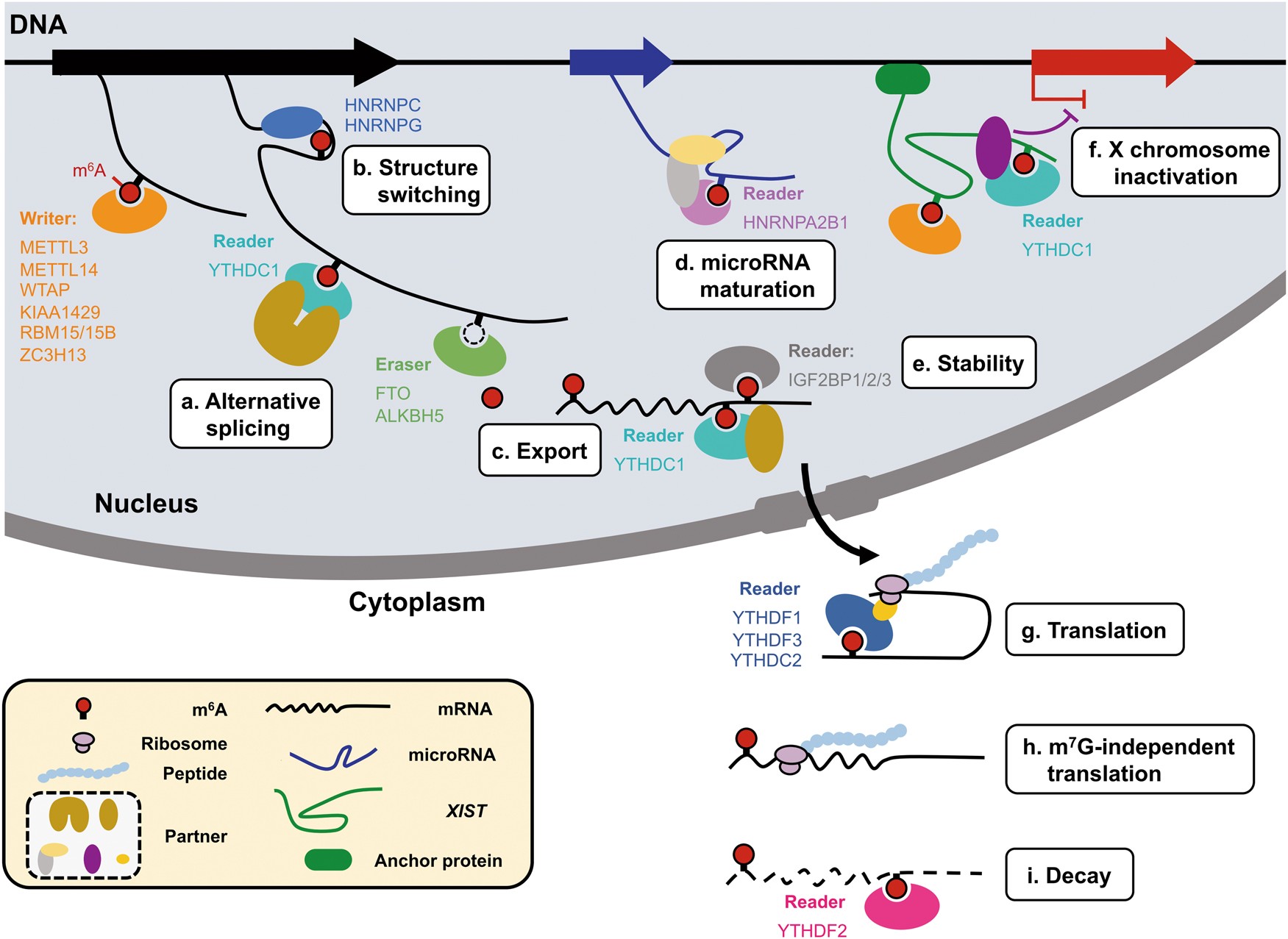
Dynamic transcriptomic m6A decoration: writers, erasers, readers and functions in RNA metabolism | Cell Research

Down-Regulation of m6A mRNA Methylation Is Involved in Dopaminergic Neuronal Death | ACS Chemical Neuroscience

Schematic diagram of miCLIP technology. m 6 A residues can be mapped by... | Download Scientific Diagram
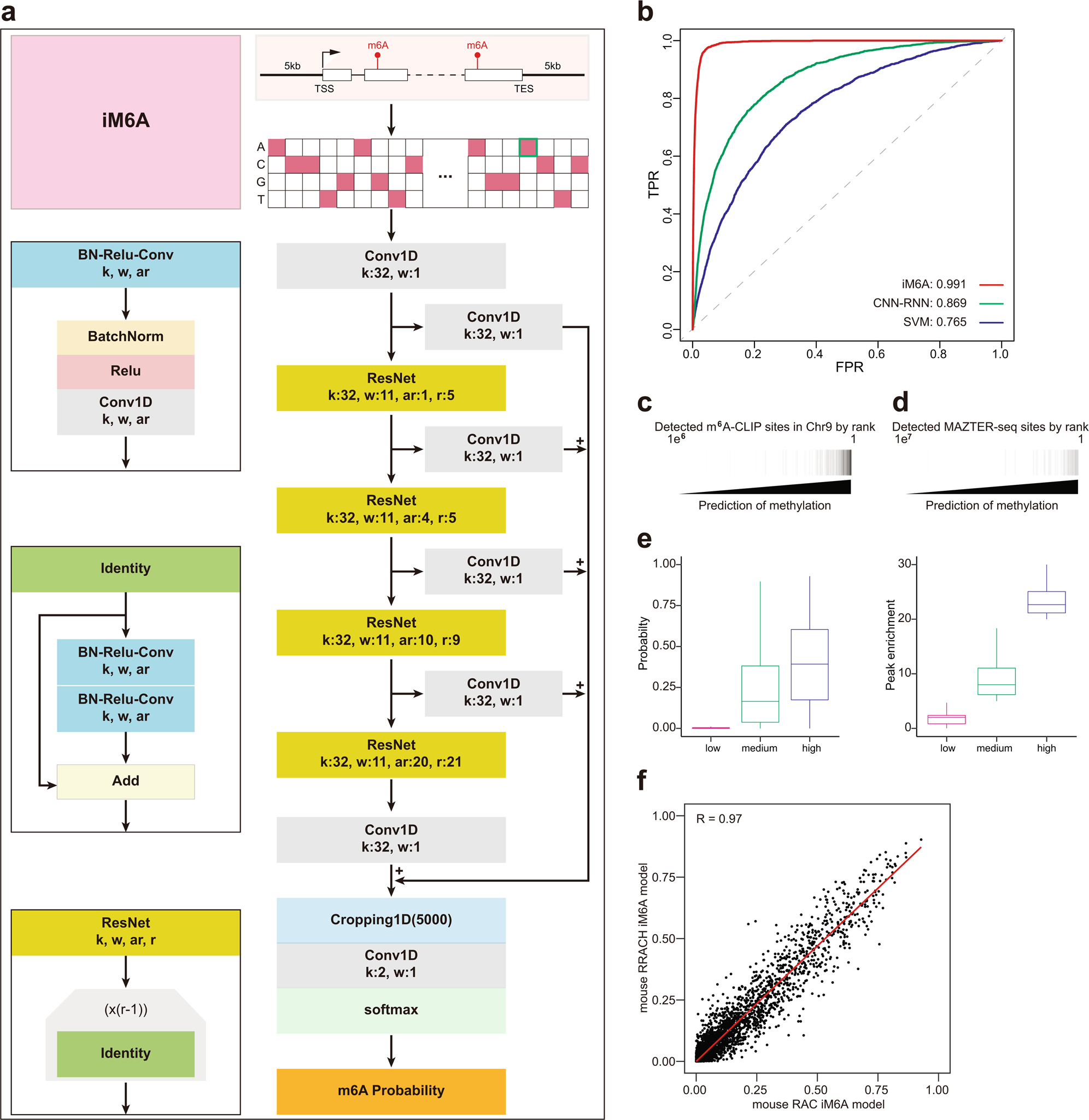
Deep learning modeling m6A deposition reveals the importance of downstream cis-element sequences | Nature Communications

Acute depletion of METTL3 identifies a role for N6-methyladenosine in alternative intron/exon inclusion in the nascent transcriptome | bioRxiv

The detection and functions of RNA modification m6A based on m6A writers and erasers - ScienceDirect
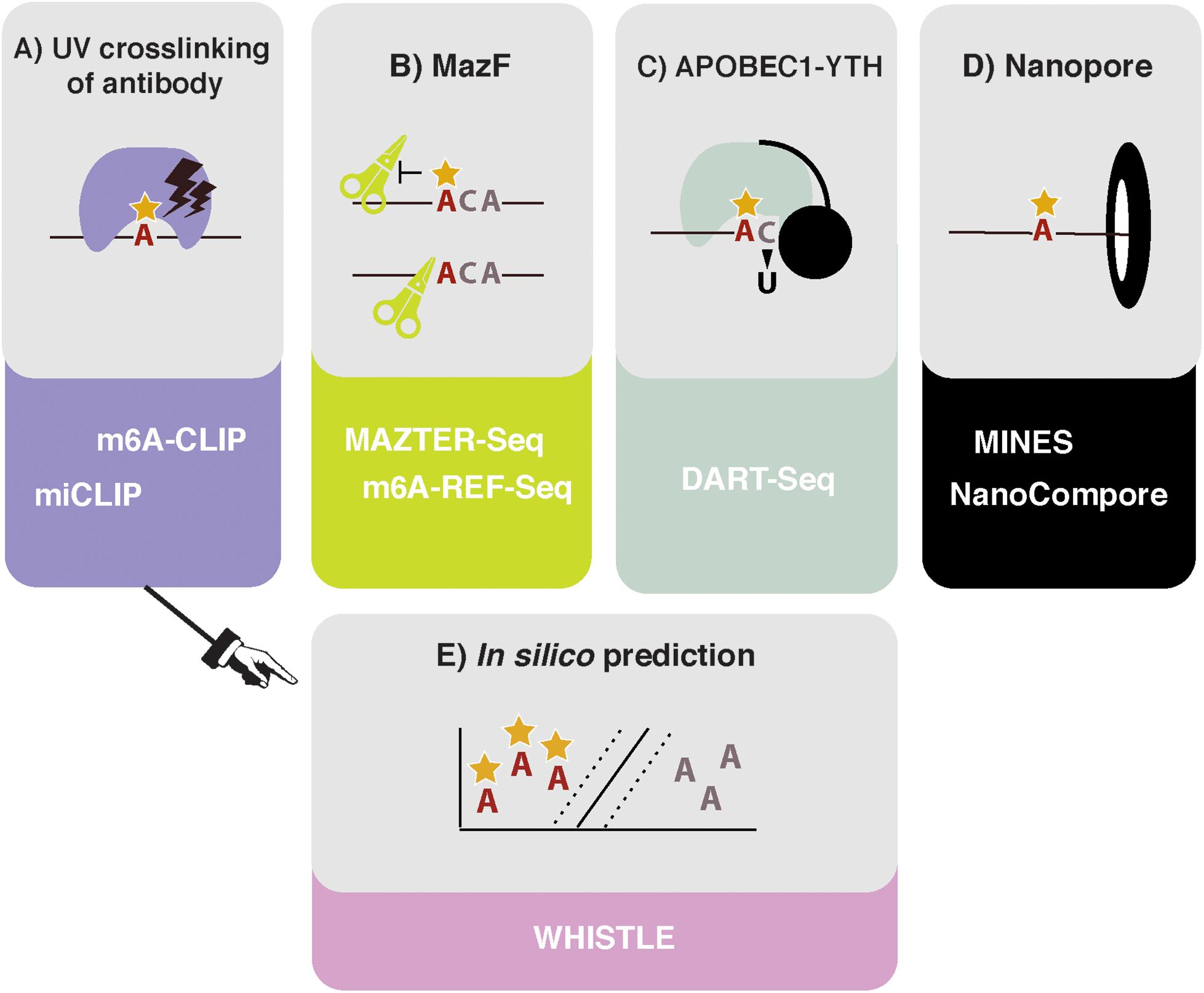
Frontiers | How Do You Identify m6 A Methylation in Transcriptomes at High Resolution? A Comparison of Recent Datasets

Antibody-free enzyme-assisted chemical approach for detection of N6-methyladenosine | Nature Chemical Biology

Relevance of N6-methyladenosine regulators for transcriptome: Implications for development and the cardiovascular system - Journal of Molecular and Cellular Cardiology
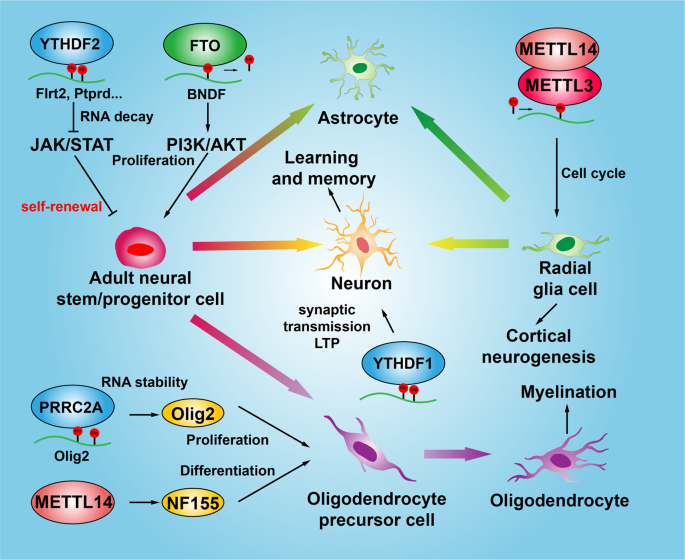
The role of m6A modification in the biological functions and diseases | Signal Transduction and Targeted Therapy